Molecular evolution
| Part of a series on |
| Evolutionary biology |
|---|
 |
Molecular evolution describes how inherited DNA and/or RNA change over evolutionary time, and the consequences of this for proteins and other components of cells and organisms. Molecular evolution is the basis of phylogenetic approaches to describing the tree of life. Molecular evolution overlaps with population genetics, especially on shorter timescales. Topics in molecular evolution include the origins of new genes, the genetic nature of complex traits, the genetic basis of adaptation and speciation, the evolution of development, and patterns and processes underlying genomic changes during evolution.
History
[edit]The history of molecular evolution starts in the early 20th century with comparative biochemistry, and the use of "fingerprinting" methods such as immune assays, gel electrophoresis, and paper chromatography in the 1950s to explore homologous proteins.[1][2] The advent of protein sequencing allowed molecular biologists to create phylogenies based on sequence comparison, and to use the differences between homologous sequences as a molecular clock to estimate the time since the most recent common ancestor.[3][1] The surprisingly large amount of molecular divergence within and between species inspired the neutral theory of molecular evolution in the late 1960s.[4][5][6] Neutral theory also provided a theoretical basis for the molecular clock, although this is not needed for the clock's validity. After the 1970s, nucleic acid sequencing allowed molecular evolution to reach beyond proteins to highly conserved ribosomal RNA sequences, the foundation of a reconceptualization of the early history of life.[1] The Society for Molecular Biology and Evolution was founded in 1982.
Molecular phylogenetics
[edit]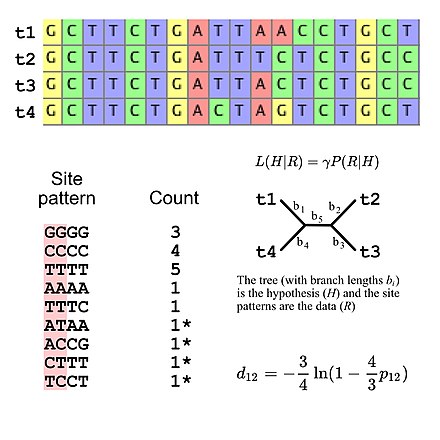
Molecular phylogenetics uses DNA, RNA, or protein sequences to resolve questions in systematics, i.e. about their correct scientific classification from the point of view of evolutionary history. The result of a molecular phylogenetic analysis is expressed in a phylogenetic tree. Phylogenetic inference is conducted using data from DNA sequencing. This is aligned to identify which sites are homologous. A substitution model describes what patterns are expected to be common or rare. Sophisticated computational inference is then used to generate one or more plausible trees.
Some phylogenetic methods account for variation among sites and among tree branches. Different genes, e.g. hemoglobin vs. cytochrome c, generally evolve at different rates.[7] These rates are relatively constant over time (e.g., hemoglobin does not evolve at the same rate as cytochrome c, but hemoglobins from humans, mice, etc. do have comparable rates of evolution), although rapid evolution along one branch can indicate increased directional selection on that branch.[8] Purifying selection causes functionally important regions to evolve more slowly, and amino acid substitutions involving similar amino acids occurs more often than dissimilar substitutions.[7]

Gene family evolution
[edit]
Gene duplication can produce multiple homologous proteins (paralogs) within the same species. Phylogenetic analysis of proteins has revealed how proteins evolve and change their structure and function over time.[9][10]
For example, ribonucleotide reductase (RNR) has evolved a multitude of structural and functional variants. Class I RNRs use a ferritin subunit and differ by the metal they use as cofactors. In class II RNRs, the thiyl radical is generated using an adenosylcobalamin cofactor and these enzymes do not require additional subunits (as opposed to class I which do). In class III RNRs, the thiyl radical is generated using S-adenosylmethionine bound to a [4Fe-4S] cluster. That is, within a single family of proteins numerous structural and functional mechanisms can evolve.[11]
In a proof-of-concept study, Bhattacharya and colleagues converted myoglobin, a non-enzymatic oxygen storage protein, into a highly efficient Kemp eliminase using only three mutations. This demonstrates that only few mutations are needed to radically change the function of a protein.[12] Directed evolution is the attempt to engineer proteins using methods inspired by molecular evolution.
Molecular evolution at one site
[edit]Change at one locus begins with a new mutation, which might become fixed due to some combination of natural selection, genetic drift, and gene conversion.
Mutation
[edit]
Mutations are permanent, transmissible changes to the genetic material (DNA or RNA) of a cell or virus. Mutations result from errors in DNA replication during cell division and by exposure to radiation, chemicals, other environmental stressors, viruses, or transposable elements. When point mutations to just one base-pair of the DNA fall within a region coding for a protein, they are characterized by whether they are synonymous (do not change the amino acid sequence) or non-synonymous. Other types of mutations modify larger segments of DNA and can cause duplications, insertions, deletions, inversions, and translocations.[13]
The distribution of rates for diverse kinds of mutations is called the "mutation spectrum" (see App. B of [14]). Mutations of different types occur at widely varying rates. Point mutation rates for most organisms are very low, roughly 10−9 to 10−8 per site per generation,[15] though some viruses have higher mutation rates on the order of 10−6 per site per generation.[16] Transitions (A ↔ G or C ↔ T) are more common than transversions (purine (adenine or guanine)) ↔ pyrimidine (cytosine or thymine, or in RNA, uracil)).[17] Perhaps the most common type of mutation in humans is a change in the length of a short tandem repeat (e.g., the CAG repeats underlying various disease-associated mutations). Such STR mutations may occur at rates on the order of 10−3 per generation.[18]
Different frequencies of different types of mutations can play an important role in evolution via bias in the introduction of variation (arrival bias), contributing to parallelism, trends, and differences in the navigability of adaptive landscapes.[19][20] Mutation bias makes systematic or predictable contributions to parallel evolution.[14] Since the 1960s, genomic GC content has been thought to reflect mutational tendencies.[21][22] Mutational biases also contribute to codon usage bias.[23] Although such hypotheses are often associated with neutrality, recent theoretical and empirical results have established that mutational tendencies can influence both neutral and adaptive evolution via bias in the introduction of variation (arrival bias).
Selection
[edit]Selection can occur when an allele confers greater fitness, i.e. greater ability to survive or reproduce, on the average individual than carries it. A selectionist approach emphasizes e.g. that biases in codon usage are due at least in part to the ability of even weak selection to shape molecular evolution.[24]
Selection can also operate at the gene level at the expense of organismal fitness, resulting in intragenomic conflict. This is because there can be a selective advantage for selfish genetic elements in spite of a host cost. Examples of such selfish elements include transposable elements, meiotic drivers, and selfish mitochondria.
Selection can be detected using the Ka/Ks ratio, the McDonald–Kreitman test. Rapid adaptive evolution is often found for genes involved in intragenomic conflict, sexual antagonistic coevolution, and the immune system.
Genetic drift
[edit]Genetic drift is the change of allele frequencies from one generation to the next due to stochastic effects of random sampling in finite populations. These effects can accumulate until a mutation becomes fixed in a population. For neutral mutations, the rate of fixation per generation is equal to the mutation rate per replication. A relatively constant mutation rate thus produces a constant rate of change per generation (molecular clock).
Slightly deleterious mutations with a selection coefficient less than a threshold value of 1 / the effective population size can also fix. Many genomic features have been ascribed to accumulation of nearly neutral detrimental mutations as a result of small effective population sizes.[25] With a smaller effective population size, a larger variety of mutations will behave as if they are neutral due to inefficiency of selection.
Gene conversion
[edit]Gene conversion occurs during recombination, when nucleotide damage is repaired using an homologous genomic region as a template. It can be a biased process, i.e. one allele may have a higher probability of being the donor than the other in a gene conversion event. In particular, GC-biased gene conversion tends to increase the GC-content of genomes, particularly in regions with higher recombination rates.[26] There is also evidence for GC bias in the mismatch repair process.[27] It is thought that this may be an adaptation to the high rate of methyl-cytosine deamination which can lead to C→T transitions.
The dynamics of biased gene conversion resemble those of natural selection, in that a favored allele will tend to increase exponentially in frequency when rare.
Genome architecture
[edit]Genome size
[edit]Genome size is influenced by the amount of repetitive DNA as well as number of genes in an organism. Some organisms, such as most bacteria, Drosophila, and Arabidopsis have particularly compact genomes with little repetitive content or non-coding DNA. Other organisms, like mammals or maize, have large amounts of repetitive DNA, long introns, and substantial spacing between genes. The C-value paradox refers to the lack of correlation between organism 'complexity' and genome size. Explanations for the so-called paradox are two-fold. First, repetitive genetic elements can comprise large portions of the genome for many organisms, thereby inflating DNA content of the haploid genome. Repetitive genetic elements are often descended from transposable elements.
Secondly, the number of genes is not necessarily indicative of the number of developmental stages or tissue types in an organism. An organism with few developmental stages or tissue types may have large numbers of genes that influence non-developmental phenotypes, inflating gene content relative to developmental gene families.
Neutral explanations for genome size suggest that when population sizes are small, many mutations become nearly neutral. Hence, in small populations repetitive content and other 'junk' DNA can accumulate without placing the organism at a competitive disadvantage. There is little evidence to suggest that genome size is under strong widespread selection in multicellular eukaryotes. Genome size, independent of gene content, correlates poorly with most physiological traits and many eukaryotes, including mammals, harbor very large amounts of repetitive DNA.
However, birds likely have experienced strong selection for reduced genome size, in response to changing energetic needs for flight. Birds, unlike humans, produce nucleated red blood cells, and larger nuclei lead to lower levels of oxygen transport. Bird metabolism is far higher than that of mammals, due largely to flight, and oxygen needs are high. Hence, most birds have small, compact genomes with few repetitive elements. Indirect evidence suggests that non-avian theropod dinosaur ancestors of modern birds[28] also had reduced genome sizes, consistent with endothermy and high energetic needs for running speed. Many bacteria have also experienced selection for small genome size, as time of replication and energy consumption are so tightly correlated with fitness.
Chromosome number and organization
[edit]The ant Myrmecia pilosula has only a single pair of chromosomes[29] whereas the Adders-tongue fern Ophioglossum reticulatum has up to 1260 chromosomes.[30] The number of chromosomes in an organism's genome does not necessarily correlate with the amount of DNA in its genome. The genome-wide amount of recombination is directly controlled by the number of chromosomes, with one crossover per chromosome or per chromosome arm, depending on the species.[31]
Changes in chromosome number can play a key role in speciation, as differing chromosome numbers can serve as a barrier to reproduction in hybrids. Human chromosome 2 was created from a fusion of two chimpanzee chromosomes and still contains central telomeres as well as a vestigial second centromere. Polyploidy, especially allopolyploidy, which occurs often in plants, can also result in reproductive incompatibilities with parental species. Agrodiatus blue butterflies have diverse chromosome numbers ranging from n=10 to n=134 and additionally have one of the highest rates of speciation identified to date.[32]
Cilliate genomes house each gene in individual chromosomes.
Organelles
[edit]
In addition to the nuclear genome, endosymbiont organelles contain their own genetic material. Mitochondrial and chloroplast DNA varies across taxa, but membrane-bound proteins, especially electron transport chain constituents are most often encoded in the organelle. Chloroplasts and mitochondria are maternally inherited in most species, as the organelles must pass through the egg. In a rare departure, some species of mussels are known to inherit mitochondria from father to son.
Origins of new genes
[edit]New genes arise from several different genetic mechanisms including gene duplication, de novo gene birth, retrotransposition, chimeric gene formation, recruitment of non-coding sequence into an existing gene, and gene truncation.
Gene duplication initially leads to redundancy. However, duplicated gene sequences can mutate to develop new functions or specialize so that the new gene performs a subset of the original ancestral functions. Retrotransposition duplicates genes by copying mRNA to DNA and inserting it into the genome. Retrogenes generally insert into new genomic locations, lack introns. and sometimes develop new expression patterns and functions.
Chimeric genes form when duplication, deletion, or incomplete retrotransposition combine portions of two different coding sequences to produce a novel gene sequence. Chimeras often cause regulatory changes and can shuffle protein domains to produce novel adaptive functions.
De novo gene birth can give rise to protein-coding genes and non-coding genes from previously non-functional DNA.[33] For instance, Levine and colleagues reported the origin of five new genes in the D. melanogaster genome.[34][35] Similar de novo origin of genes has been also shown in other organisms such as yeast,[36] rice[37] and humans.[38] De novo genes may evolve from spurious transcripts that are already expressed at low levels.[39]
Constructive neutral evolution
[edit]Constructive neutral evolution (CNE) explains that complex systems can emerge and spread into a population through neutral transitions with the principles of excess capacity, presuppression, and ratcheting,[40][41][42] and it has been applied in areas ranging from the origins of the spliceosome to the complex interdependence of microbial communities.[43][44][45]
Journals and societies
[edit]The Society for Molecular Biology and Evolution publishes the journals "Molecular Biology and Evolution" and "Genome Biology and Evolution" and holds an annual international meeting. Other journals dedicated to molecular evolution include Journal of Molecular Evolution and Molecular Phylogenetics and Evolution. Research in molecular evolution is also published in journals of genetics, molecular biology, genomics, systematics, and evolutionary biology.
See also
[edit]- Evolution
- E. coli long-term evolution experiment
- Evolutionary physiology
- Genomic organization
- Genome evolution
- Heterotachy
- History of molecular evolution
- Horizontal gene transfer
- Human evolution
- Molecular clock
- Molecular paleontology
- Nearly neutral theory of molecular evolution
- Neutral theory of molecular evolution
- Nucleotide diversity
- Phylogenetic comparative methods
- Phylogenetics
- Population genetics
- Selection
References
[edit]- ^ a b c Dietrich MR (1998). "Paradox and persuasion: negotiating the place of molecular evolution within evolutionary biology". Journal of the History of Biology. 31 (1): 85–111. doi:10.1023/A:1004257523100. PMID 11619919. S2CID 29935487.
- ^ Hagen JB (1999). "Naturalists, molecular biologists, and the challenges of molecular evolution". Journal of the History of Biology. 32 (2): 321–341. doi:10.1023/A:1004660202226. PMID 11624208. S2CID 26994015.
- ^ Zuckerkandl, Emile; Pauling, Linus (March 1965). "Molecules as documents of evolutionary history". Journal of Theoretical Biology. 8 (2): 357–366. Bibcode:1965JThBi...8..357Z. doi:10.1016/0022-5193(65)90083-4. PMID 5876245.
- ^ Kimura M (February 1968). "Evolutionary rate at the molecular level". Nature. 217 (5129): 624–626. Bibcode:1968Natur.217..624K. doi:10.1038/217624a0. PMID 5637732. S2CID 4161261.
- ^ King JL, Jukes TH (May 1969). "Non-Darwinian evolution". Science. 164 (3881): 788–798. Bibcode:1969Sci...164..788L. doi:10.1126/science.164.3881.788. PMID 5767777.
- ^ Kimura, M. (1983). The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge. ISBN 0-521-23109-4.
- ^ a b Fay JC, Wu CI (2003). "Sequence divergence, functional constraint, and selection in protein evolution". Annual Review of Genomics and Human Genetics. 4: 213–235. doi:10.1146/annurev.genom.4.020303.162528. PMID 14527302. S2CID 6360375.
- ^ Álvarez-Carretero, Sandra; Kapli, Paschalia; Yang, Ziheng (4 April 2023). "Beginner's Guide on the Use of PAML to Detect Positive Selection". Molecular Biology and Evolution. 40 (4). doi:10.1093/molbev/msad041. PMC 10127084. PMID 37096789.
- ^ Hanukoglu I (February 2017). "ASIC and ENaC type sodium channels: conformational states and the structures of the ion selectivity filters". The FEBS Journal. 284 (4): 525–545. doi:10.1111/febs.13840. PMID 27580245. S2CID 24402104.
- ^ Hanukoglu I, Hanukoglu A (April 2016). "Epithelial sodium channel (ENaC) family: Phylogeny, structure-function, tissue distribution, and associated inherited diseases". Gene. 579 (2): 95–132. doi:10.1016/j.gene.2015.12.061. PMC 4756657. PMID 26772908.
- ^ Burnim AA, Spence MA, Xu D, Jackson CJ, Ando N (September 2022). Ben-Tal N, Weigel D, Ben-Tal N, Stubbe J, Hofer A (eds.). "Comprehensive phylogenetic analysis of the ribonucleotide reductase family reveals an ancestral clade". eLife. 11: e79790. doi:10.7554/eLife.79790. PMC 9531940. PMID 36047668.
- ^ Bhattacharya S, Margheritis EG, Takahashi K, Kulesha A, D'Souza A, Kim I, et al. (October 2022). "NMR-guided directed evolution". Nature. 610 (7931): 389–393. Bibcode:2022Natur.610..389B. doi:10.1038/s41586-022-05278-9. PMC 10116341. PMID 36198791. S2CID 245067145.
- ^ Yang, J. (2016, March 23). What are Genetic Mutation? Retrieved from https://www.singerinstruments.com/resource/what-are-genetic-mutation/ .
- ^ a b A. Stoltzfus (2021). Mutation, Randomness and Evolution. Oxford, Oxford.
- ^ Wang, Yiguan; Obbard, Darren J (19 July 2023). "Experimental estimates of germline mutation rate in eukaryotes: a phylogenetic meta-analysis". Evolution Letters. 7 (4): 216–226. doi:10.1093/evlett/qrad027. hdl:20.500.11820/8ffd5b76-77ae-4764-ae31-de2fb8aa35cf. PMC 10355183. PMID 37475753.
- ^ Peck, Kayla M.; Lauring, Adam S. (15 July 2018). "Complexities of Viral Mutation Rates". Journal of Virology. 92 (14): e01031-17. doi:10.1128/JVI.01031-17. PMC 6026756. PMID 29720522.
- ^ "Transitions vs transversions".
- ^ J. L. Weber and C. Wong (1993). "Mutation of human short tandem repeats". Hum Mol Genet. 2 (8): 1123–8. doi:10.1093/hmg/2.8.1123. PMID 8401493.
- ^ A. V. Cano and J. L. Payne (2020). "Mutation bias interacts with composition bias to influence adaptive evolution". PLOS Computational Biology. 16 (9): e1008296. Bibcode:2020PLSCB..16E8296C. doi:10.1371/journal.pcbi.1008296. PMC 7571706. PMID 32986712.
- ^ M. Nei (2013). Mutation-Driven Evolution. Oxford University Press.
- ^ E. Freese (1962). "On the Evolution of the Base Composition of DNA". J. Theor. Biol. 3 (1): 82–101. Bibcode:1962JThBi...3...82F. doi:10.1016/S0022-5193(62)80005-8.
It is unimportant in this connection whether selection has been negligible or self-cancelling.
- ^ N. Sueoka (1962). "On the Genetic Basis of Variation and Heterogeneity of DNA Base Composition". Proc. Natl. Acad. Sci. U.S.A. 48 (4): 582–592. Bibcode:1962PNAS...48..582S. doi:10.1073/pnas.48.4.582. PMC 220819. PMID 13918161.
- ^ A. Stoltzfus and L. Y. Yampolsky (2009). "Climbing mount probable: mutation as a cause of nonrandomness in evolution". J Hered. 100 (5): 637–47. doi:10.1093/jhered/esp048. PMID 19625453.
- ^ Hershberg R, Petrov DA (December 2008). "Selection on codon bias". Annual Review of Genetics. 42 (1): 287–299. doi:10.1146/annurev.genet.42.110807.091442. PMID 18983258. S2CID 7085012.
- ^ Lynch M (2007). The Origins of Genome Architecture. Sinauer. ISBN 978-0-87893-484-3.
- ^ Duret L, Galtier N (2009). "Biased gene conversion and the evolution of mammalian genomic landscapes". Annual Review of Genomics and Human Genetics. 10: 285–311. doi:10.1146/annurev-genom-082908-150001. PMID 19630562.
- ^ Galtier N, Piganeau G, Mouchiroud D, Duret L (October 2001). "GC-content evolution in mammalian genomes: the biased gene conversion hypothesis". Genetics. 159 (2): 907–911. doi:10.1093/genetics/159.2.907. PMC 1461818. PMID 11693127.
- ^ Organ CL, Shedlock AM, Meade A, Pagel M, Edwards SV (March 2007). "Origin of avian genome size and structure in non-avian dinosaurs". Nature. 446 (7132): 180–184. Bibcode:2007Natur.446..180O. doi:10.1038/nature05621. PMID 17344851. S2CID 3031794.
- ^ Crosland MW, Crozier RH (March 1986). "Myrmecia pilosula, an Ant with Only One Pair of Chromosomes". Science. 231 (4743): 1278. Bibcode:1986Sci...231.1278C. doi:10.1126/science.231.4743.1278. PMID 17839565. S2CID 25465053.
- ^ Gerardus J. H. Grubben (2004). Vegetables. PROTA. p. 404. ISBN 978-90-5782-147-9. Retrieved 10 March 2013.
- ^ Pardo-Manuel de Villena, Fernando; Sapienza, Carmen (April 2001). "Recombination is proportional to the number of chromosome arms in mammals". Mammalian Genome. 12 (4): 318–322. doi:10.1007/s003350020005. PMID 11309665.
- ^ Kandul NP, Lukhtanov VA, Pierce NE (March 2007). "Karyotypic diversity and speciation in Agrodiaetus butterflies". Evolution; International Journal of Organic Evolution. 61 (3): 546–559. doi:10.1111/j.1558-5646.2007.00046.x. PMID 17348919.
- ^ McLysaght A, Guerzoni D (September 2015). "New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 370 (1678): 20140332. doi:10.1098/rstb.2014.0332. PMC 4571571. PMID 26323763.
- ^ Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ (June 2006). "Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression". Proceedings of the National Academy of Sciences of the United States of America. 103 (26): 9935–9939. Bibcode:2006PNAS..103.9935L. doi:10.1073/pnas.0509809103. PMC 1502557. PMID 16777968.
- ^ Zhou Q, Zhang G, Zhang Y, Xu S, Zhao R, Zhan Z, et al. (September 2008). "On the origin of new genes in Drosophila". Genome Research. 18 (9): 1446–1455. doi:10.1101/gr.076588.108. PMC 2527705. PMID 18550802.
- ^ Cai J, Zhao R, Jiang H, Wang W (May 2008). "De novo origination of a new protein-coding gene in Saccharomyces cerevisiae". Genetics. 179 (1): 487–496. doi:10.1534/genetics.107.084491. PMC 2390625. PMID 18493065.
- ^ Xiao W, Liu H, Li Y, Li X, Xu C, Long M, Wang S (2009). El-Shemy HA (ed.). "A rice gene of de novo origin negatively regulates pathogen-induced defense response". PLOS ONE. 4 (2): e4603. Bibcode:2009PLoSO...4.4603X. doi:10.1371/journal.pone.0004603. PMC 2643483. PMID 19240804.
- ^ Knowles DG, McLysaght A (October 2009). "Recent de novo origin of human protein-coding genes". Genome Research. 19 (10): 1752–1759. doi:10.1101/gr.095026.109. PMC 2765279. PMID 19726446.
- ^ Wilson BA, Masel J (2011). "Putatively noncoding transcripts show extensive association with ribosomes". Genome Biology and Evolution. 3: 1245–1252. doi:10.1093/gbe/evr099. PMC 3209793. PMID 21948395.
- ^ Stoltzfus A (August 1999). "On the possibility of constructive neutral evolution". Journal of Molecular Evolution. 49 (2): 169–181. Bibcode:1999JMolE..49..169S. doi:10.1007/PL00006540. PMID 10441669. S2CID 1743092.
- ^ Stoltzfus A (October 2012). "Constructive neutral evolution: exploring evolutionary theory's curious disconnect". Biology Direct. 7 (1): 35. doi:10.1186/1745-6150-7-35. PMC 3534586. PMID 23062217.
- ^ Muñoz-Gómez SA, Bilolikar G, Wideman JG, Geiler-Samerotte K (April 2021). "Constructive Neutral Evolution 20 Years Later". Journal of Molecular Evolution. 89 (3): 172–182. Bibcode:2021JMolE..89..172M. doi:10.1007/s00239-021-09996-y. PMC 7982386. PMID 33604782.
- ^ Lukeš J, Archibald JM, Keeling PJ, Doolittle WF, Gray MW (July 2011). "How a neutral evolutionary ratchet can build cellular complexity". IUBMB Life. 63 (7): 528–537. doi:10.1002/iub.489. PMID 21698757. S2CID 7306575.
- ^ Vosseberg J, Snel B (December 2017). "Domestication of self-splicing introns during eukaryogenesis: the rise of the complex spliceosomal machinery". Biology Direct. 12 (1): 30. doi:10.1186/s13062-017-0201-6. PMC 5709842. PMID 29191215.
- ^ Brunet TD, Doolittle WF (19 March 2018). "The generality of Constructive Neutral Evolution". Biology & Philosophy. 33 (1): 2. doi:10.1007/s10539-018-9614-6. ISSN 1572-8404. S2CID 90290787.
Further reading
[edit]- Li WH (2006). Molecular Evolution. Sinauer. ISBN 0-87893-480-4.
- Lynch M (2007). The Origins of Genome Architecture. Sinauer. ISBN 978-0-87893-484-3.
- Meyer A, van de Peer Y, eds. (2003). Genome evolution : gene and genome duplications and the origin of novel gene functions. Dordrecht: Kluwer Academic Pub. ISBN 978-1-4020-1021-7.
- Gregory TR (2005). The evolution of the genome. Burlington, MA: Elsevier Academic. ISBN 978-0-12-301463-4.
- Levinson G (2020). Rethinking evolution : the revolution that's hiding in plain sight. London: World Scientific. ISBN 978-1-78634-726-8.
- Graur D, Li WH (2000). Fundamentals of molecular evolution. Sinauer. ISBN 0-87893-266-6.
- Graur D (2016). Molecular and genome evolution. Sunderland (Mass.): Sinauer associates, Inc. ISBN 978-1605354699.
Category: molecular evolution (kimura 1968)
